Mastering Lineplots_LB
master-lineplots.RmdIn this vingette we will see how to master the Lineplots_LB function to build perfect lineplots. First we configure the environment and load all necessary packages.
After that we can load the dataframe from which we will run the lineplots.
set.seed(2201)
data01 <- data.frame(ID = rep(paste0("PT-", 1:6), each = 5),
Time = rep(1:5, 6),
Marker_1 = rnorm(30),
Marker_2 = rnorm(30, 0, 5),
Marker_3 = rnorm(30, 0, 2),
Gender = factor(rep(c("M", "F"), each = 5, length.out = 30),
levels = c("M", "F"), ordered = TRUE))In this case the generated data is simulated, for more info on how to set a dataframe compliant with the LandS package consult the “Getting Started” vignette
Single graph
You can build a single graph using the following code:
Lineplots_LB(data = data01,
variables = "Marker_1",
time = "Time")
#> Loading required package: ggpubr
#> Loading required package: dplyr
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
#> Loading required package: ggh4x
#> Loading required package: grid
#> Loading required package: gridExtra
#>
#> Attaching package: 'gridExtra'
#> The following object is masked from 'package:dplyr':
#>
#> combine
#> Loading required package: svMisc
#>
#> Attaching package: 'svMisc'
#> The following object is masked from 'package:utils':
#>
#> ?
#> Loading required package: officer
#> Loading required package: rvg
#> Loading required package: progress
#>
#> Attaching package: 'pryr'
#> The following object is masked from 'package:dplyr':
#>
#> where
#> ████████████████████████████████████████████████████████████████████████████████ 100% 1/1 | 00:00:00 | ETA: 00:00:00 | RAM: 0.07Gb
#> [[1]]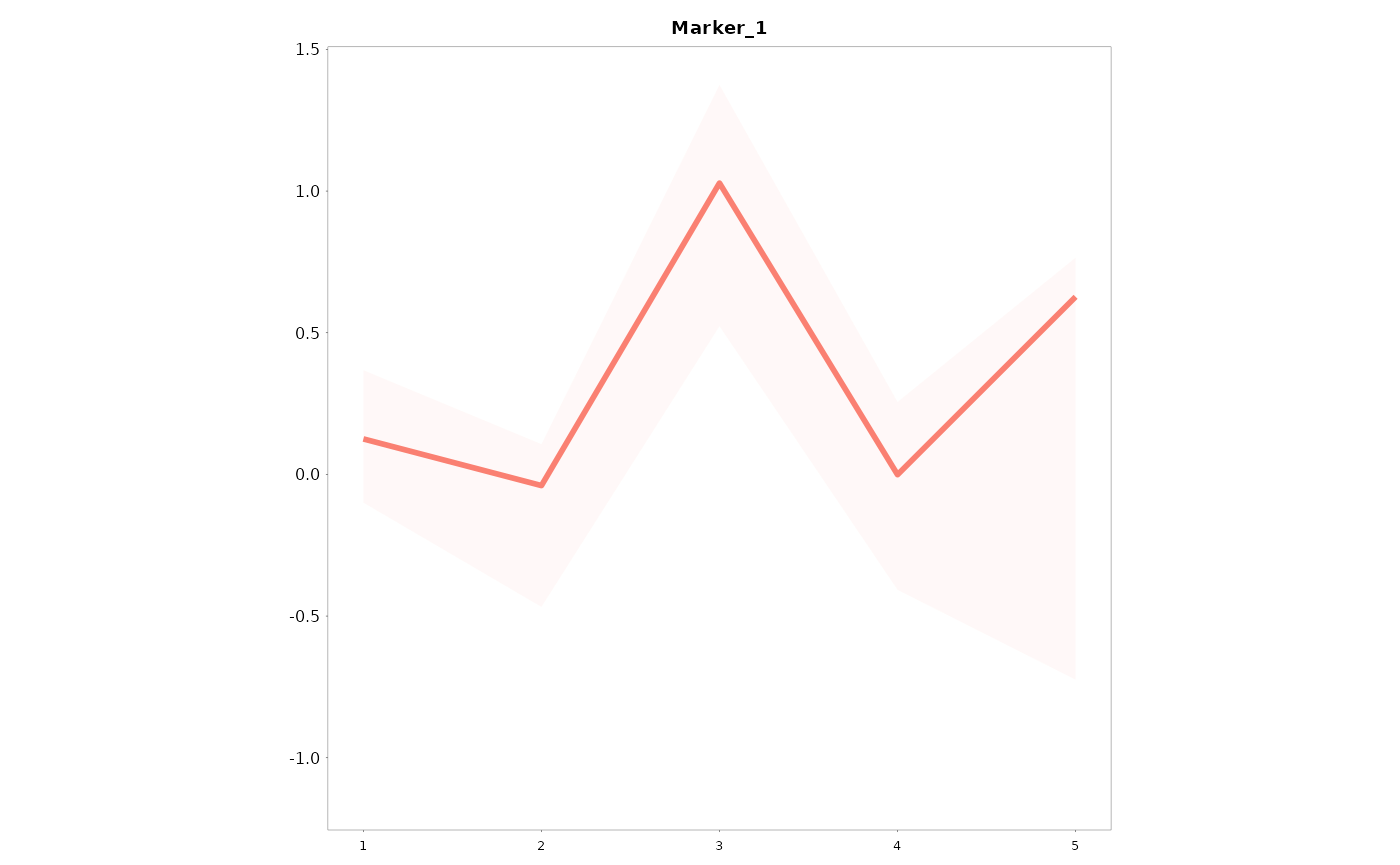
You can see that the arguments used by the function are:
data, the dataframe
variable, the variable to build the lineplots
time, the x-axis variable
Customize a single graph
We can change the lineplot graphics as follows:
Lineplots_LB(data = data01, variables = "Marker_1", time = "Time")
#> ████████████████████████████████████████████████████████████████████████████████ 100% 1/1 | 00:00:00 | ETA: 00:00:00 | RAM: 0.08Gb
#> [[1]]
Single graph split by a group variable
We can also get separated curves for a specific grouping variable:
Lineplots_LB(data = data01, variables = "Marker_1", time = "Time", group = "Gender")
#> ████████████████████████████████████████████████████████████████████████████████ 100% 1/1 | 00:00:00 | ETA: 00:00:00 | RAM: 0.08Gb
#> [[1]]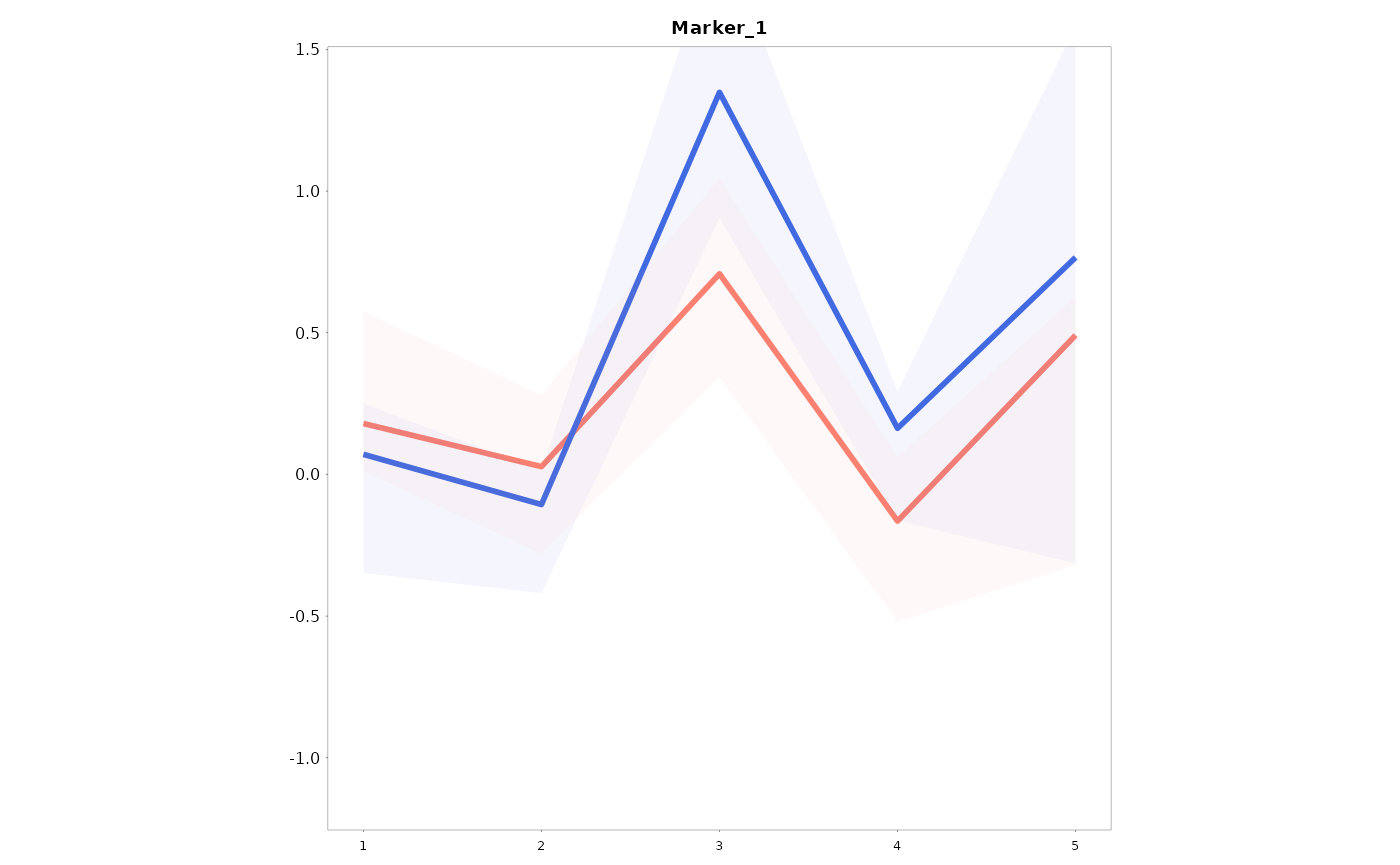
Build a list of lineplots
Lineplots_LB(data = data01, variables = c("Marker_1", "Marker_2"), time = "Time", group = "Gender")
#> ████████████████████████████████████████░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░ 50% 1/2 | 00:00:00 | ETA: 00:00:00 | RAM: 0.08Gb████████████████████████████████████████████████████████████████████████████████ 100% 2/2 | 00:00:00 | ETA: 00:00:00 | RAM: 0.08Gb
#> [[1]]
#>
#> [[2]]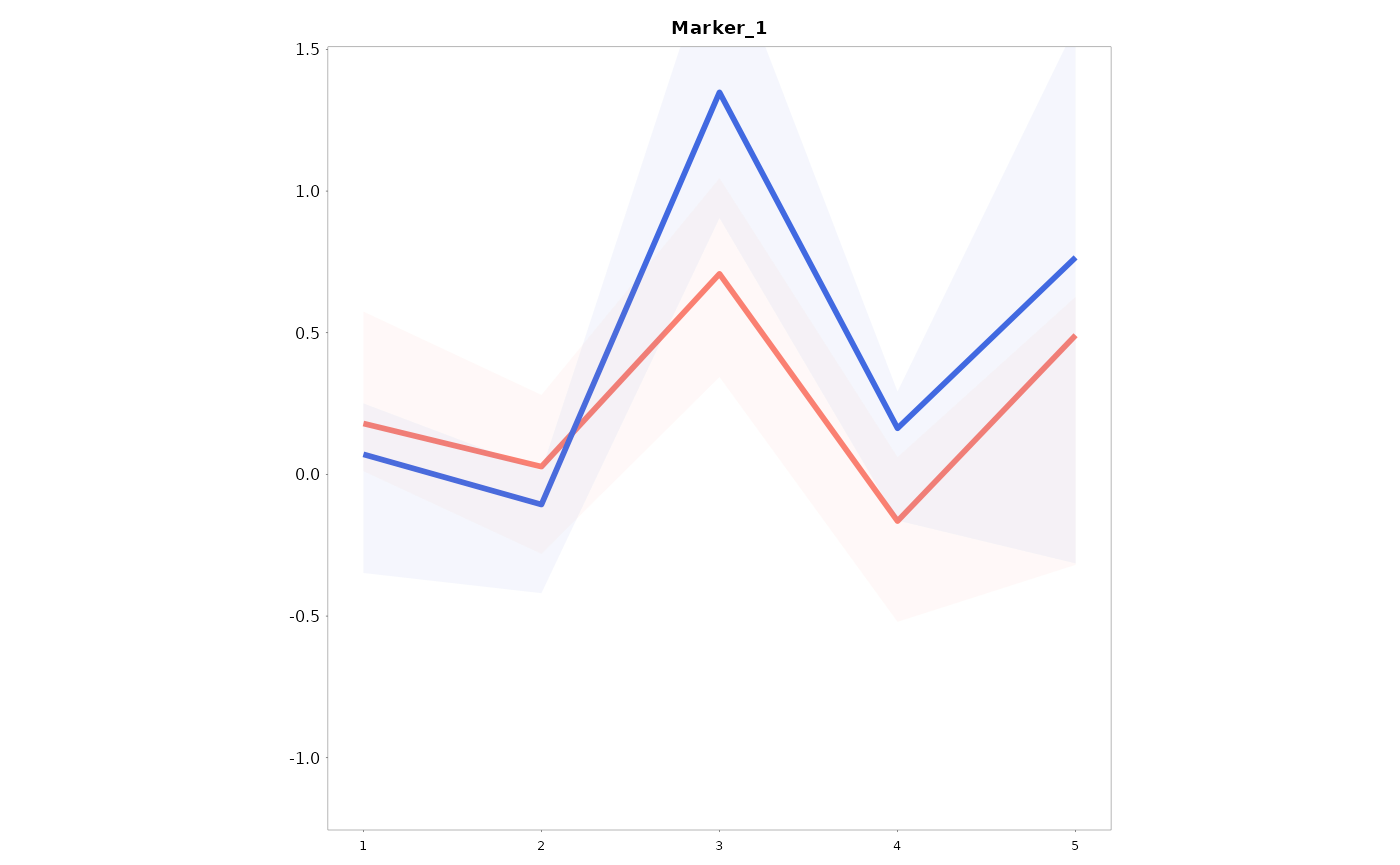
#>
#> [[3]]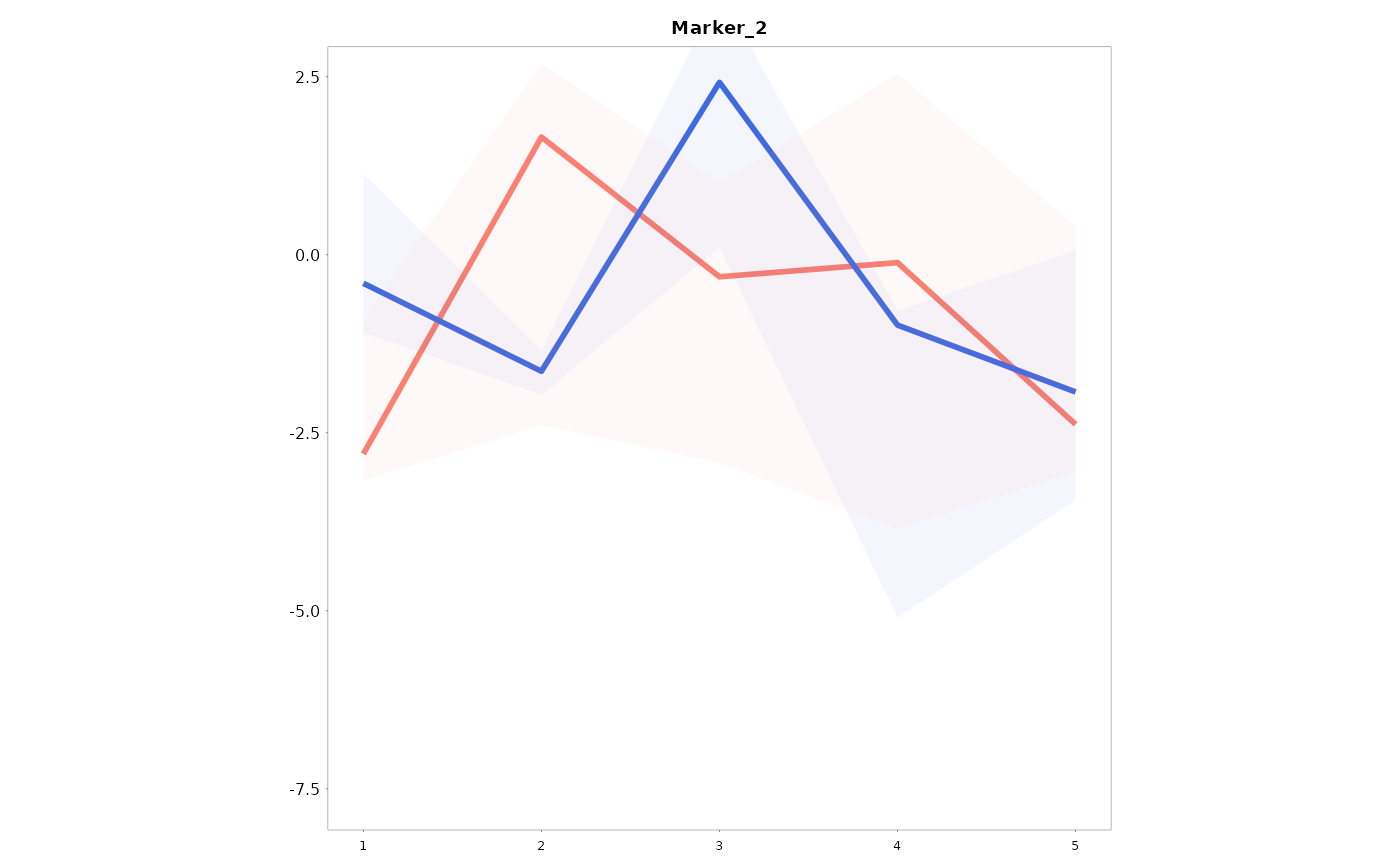
Save a list of lineplots
You can save a list of lineplots in two different ways: you can build a grid and save the entire grid in a pdf file, or you can generate a PPTX file with every lineplot in a different slide.
Save a list of lineplots as a pdf grid file
List_reg <- Lineplots_LB(data = data01, variables = c("Marker_1", "Marker_2"), time = "Time", group = "Gender")
LandS::Print_LB(plot_list = List_reg, path_print = "your/directory", ext = "pdf")Save a list of lineplots as a PPTX file
Lineplots_LB(data = data01, variables = c("Marker_1", "Marker_2"), time = "Time", group = "Gender",
PPTX = TRUE, grid = FALSE, pptx_width = 6, pptx_height = 4, target = "your/directory/new_presentation.pptx")Customize the colour of the title
library(dplyr)
df_legend <- matrix(ncol = 2, nrow = 2) %>% as.data.frame() %>% `colnames<-`(c("Var", "Colour"))
df_legend$Var <- c("Marker_1", "Marker_2")
df_legend$Colour <- c("red", "orange")
colour_title <- function(i) df_legend$Colour[df_legend$Var == i]
Lineplots_LB(data = data01, variables = c("Marker_1", "Marker_2"), time = "Time", group = "Gender",
col_title = TRUE, colour_title = colour_title)
#> ████████████████████████████████████████░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░ 50% 1/2 | 00:00:00 | ETA: 00:00:00 | RAM: 0.08Gb████████████████████████████████████████████████████████████████████████████████ 100% 2/2 | 00:00:00 | ETA: 00:00:00 | RAM: 0.08Gb
#> [[1]]
#>
#> [[2]]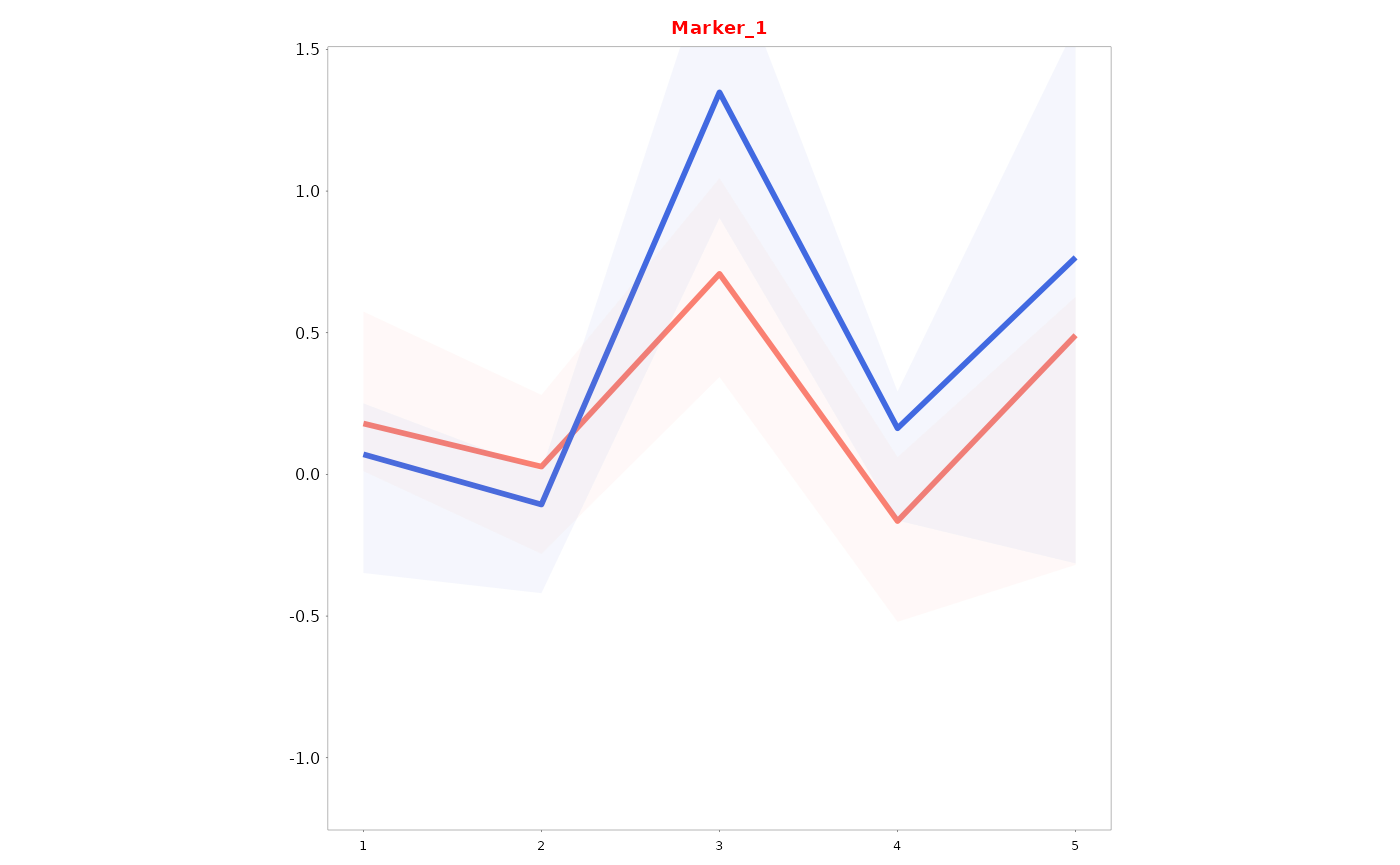
#>
#> [[3]]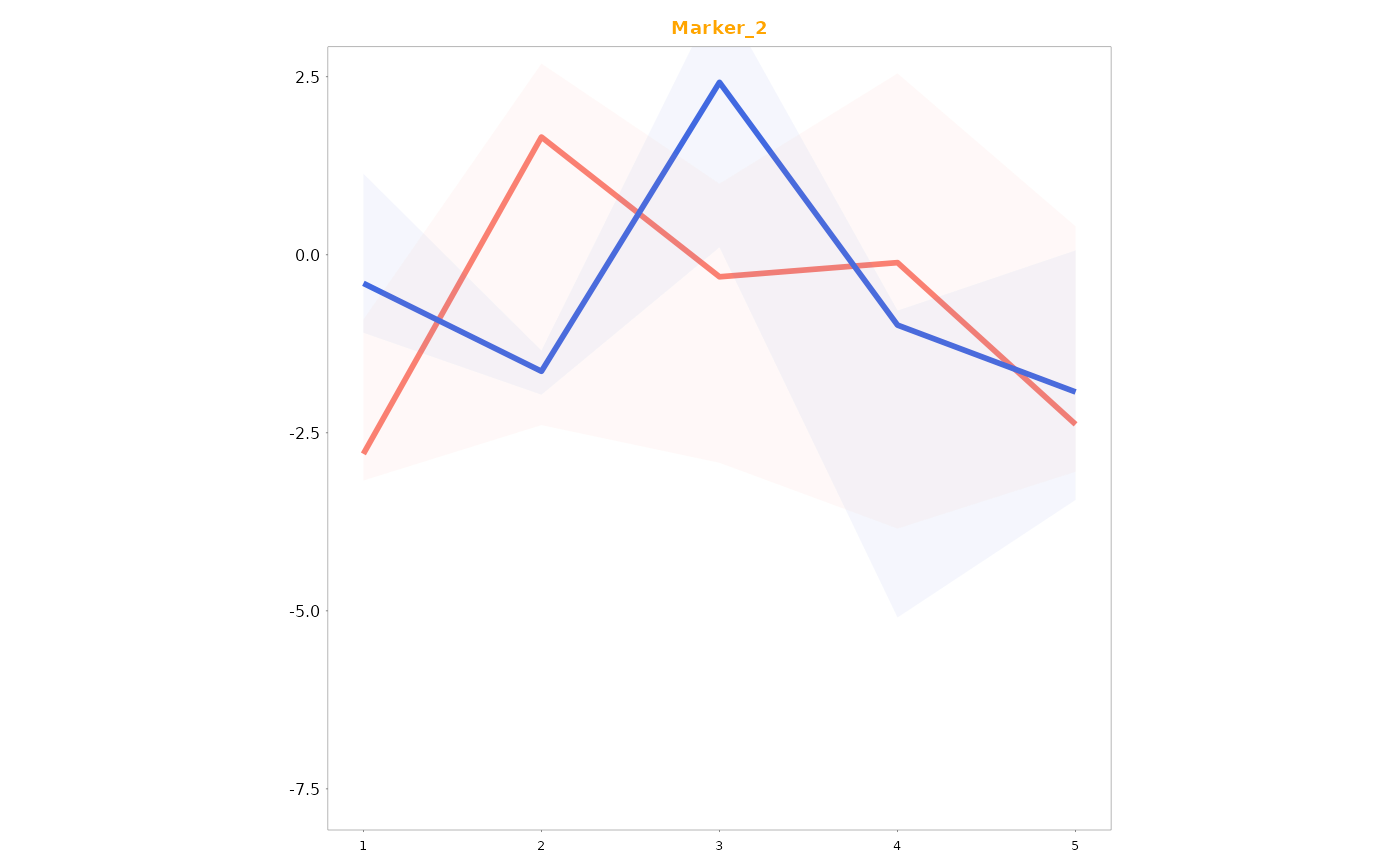
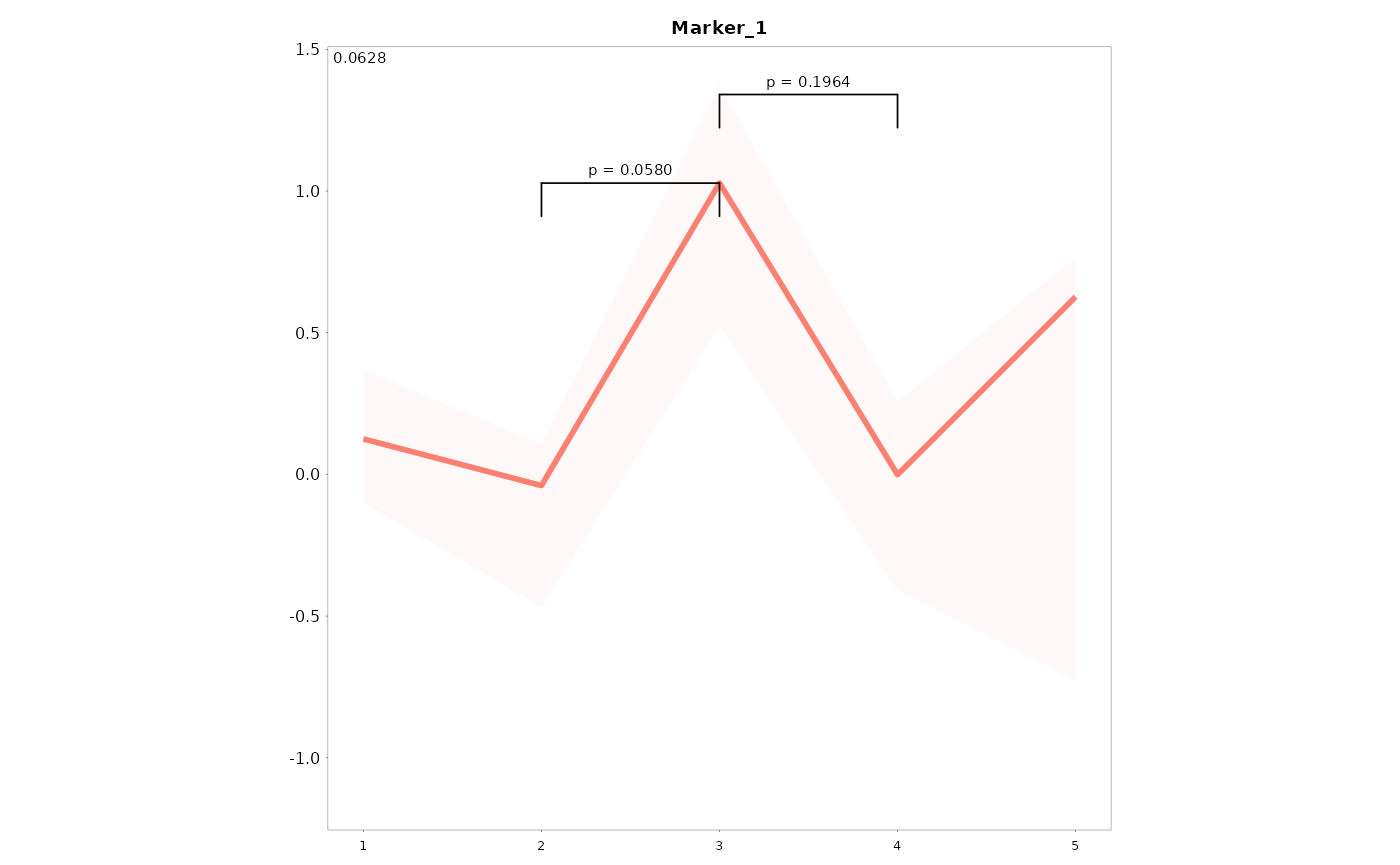
Add overall and posthoc tests to the graphs
# Let's make the grouping variable a factor
data01$Time_fac <- factor(data01$Time, levels = c(1:5), ordered = TRUE)
# Run cont_var_test
Tests <- LandS::cont_var_test_LB(data = data01, variables = c("Marker_1", "Marker_2"), group = "Time_fac",
paired = TRUE, p.adjust.method = "bonferroni")
#> Loading required package: PMCMRplus
#> Loading required package: rlang
#>
#> Attaching package: 'rlang'
#> The following object is masked from 'package:pryr':
#>
#> bytes
#> Loading required package: gtsummary
#> ████████████████████████████████████████░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░ 50% 1/2 | 00:00:00 | ETA: 00:00:00 | RAM: 0.10Gb████████████████████████████████████████████████████████████████████████████████ 100% 2/2 | 00:00:00 | ETA: 00:00:00 | RAM: 0.10Gb
#> Friedman rank sum test used
Lineplots_LB(data = data01, time = "Time", variables = "Marker_1", Overall = TRUE, Test_results = Tests[[3]],
Posthoc = TRUE, posthoc_test_size = 2, threshold_posthoc = 1)
#> ████████████████████████████████████████████████████████████████████████████████ 100% 1/1 | 00:00:00 | ETA: 00:00:00 | RAM: 0.10Gb
#> [[1]]